Science Spotlight - pIGLETs
The Day that pIGLETs Swim: A New Tool for Reliable Zebrafish Transgenesis
By Jennifer Fiene
Recently, the creation of transgenic lines has become necessary for in vivo research. The ability to examine the effects of an exogenous gene within a working biological system is a powerful tool that has facilitated research on development, disease and physiology. Zebrafish are an excellent candidate for transgenesis due to their external development, rapid maturation, and high fecundity. Transgenic technologies have progressed quickly, and zebrafish researchers now have access to many different techniques including I-SceI meganucleases, Tol2 and Ac/Ds transposons, and CRISPR-Cas91–4. These techniques have their merits, but do not come without difficulties. Issues with these techniques include random integration location (I-SceI, Tol2), variable copy numbers (I-SceI), and inconsistency in functionality and expression levels depending on loci (I-SceI, Tol2, CRISPR-Cas9)1–4. These issues collectively result in intensive screens that require considerable time and resources to generate a single, high-quality transgenic line. These are issues that led Dr. Christian Mosimann and his team at the University of Colorado to create a new transgenesis tool for zebrafish based on phiC31 integrase, an enzyme adapted from the phiC31 bacteriophage5,6. phiC31 works by recognizing attP “receiver” sites in a genomic sequence, and directionally recombining an attB flanked “donor” sequence1,6,7. Generation of transgenic lines containing an attP sequence in the genome allows vectors containing attB sites to be integrated into that exact locus8,9. This technique has been used extensively in Drosophila and mice and has allowed researchers to take advantage of “safe-harbour” loci. Safe-harbours are genomic loci where transgenes can be integrated with minimal positional effects altering expression. Placing attP sites into safe-harbours enables efficient recombination and more consistent expression, significantly reducing the amount of founder screening needed. Validated safe-harbour attP site lines in Drosophila and mice revolutionized transgenesis, however very few such lines had been created in zebrafish1,8–11. A previous paper by Roberts et al. (2014) reported creation of attP site lines in zebrafish with a screening system using lens markers, however these lines were not rigorously tested for positional effects or multi-generation stability12. Mosimann's lab has now extensively tested and validated safe-harbour attP zebrafish lines compatible with phiC31 transgenesis.
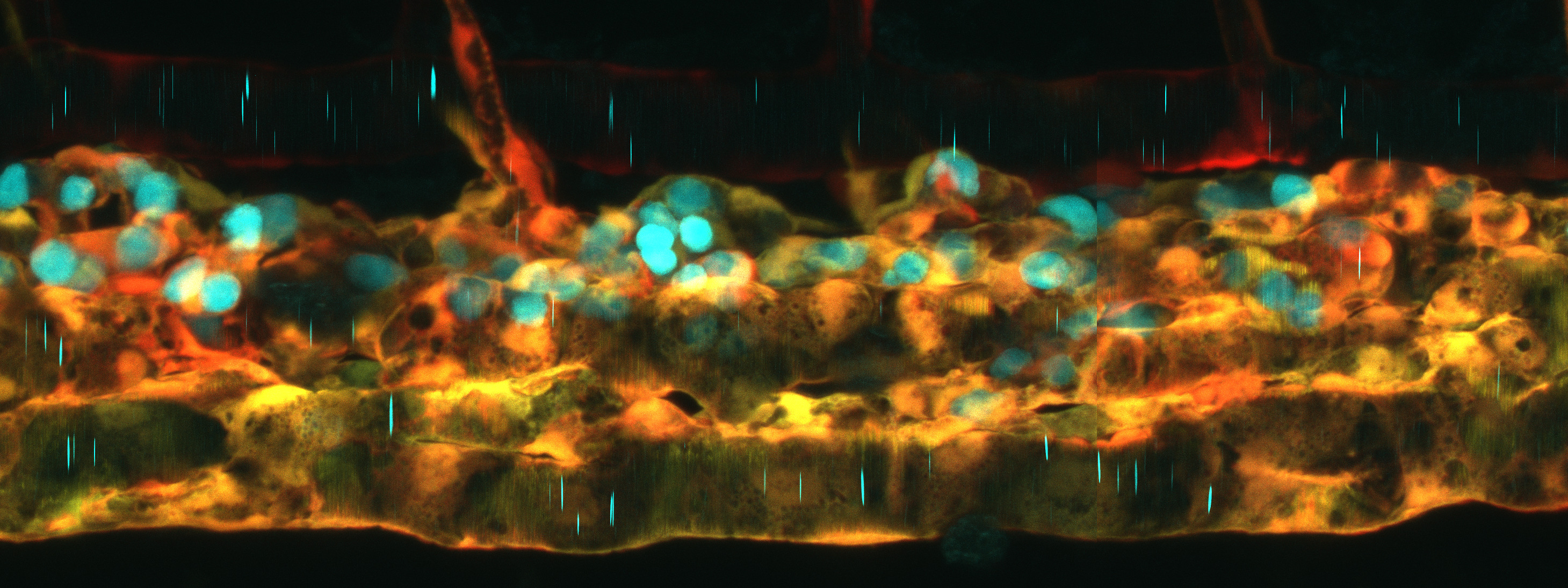
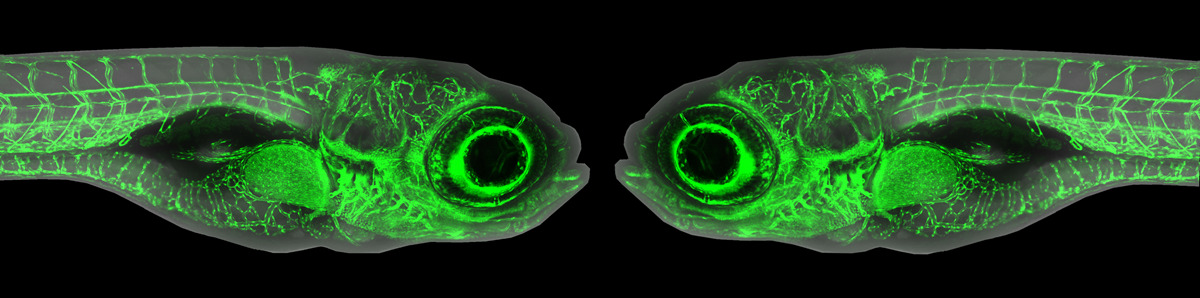
The first step of this process was to identify safe-harbours in the zebrafish genome. Luckily, Mosimann’s team had a shortcut for this: use two robust Tol2-generated transgenic lines as a guiding light to find safe-harbours. The ubi:loxP-EGFP-loxP_mCherry (ubi:Switch) and hsp70l:loxP-STOP-loxP_EGFP (hsp70l:Switch) lines both demonstrate reliable expression, dependable Cre/lox switch functionality, and are homozygous viable. All of these qualities led the authors to hypothesize that these transgenes were in safe-harbours. Using CRISPR-Cas9, Dr. Robert Lalonde, post-doctoral fellow in the Mosimann lab, was able to use a two-step process to cut out the existing transgenes and replace them with attP sites. These new attP lines were named “phiC31 Integrase Genomic Loci Engineered for Transgenesis” (pIGLET). Additionally, the team developed six complementary plasmid backbones containing attB sites that are compatible with different cloning approaches to facilitate easy vector design.
Lalonde and the Mosimann team then performed validation experiments to confirm their hunch. After injection of multiple different lineage-tracing reporter vectors into both pIGLET lines, they were able to replicate expression of well-established Tol2-based reporter lines under the same promoters. Further, it was shown that both pIGLET loci were extremely efficient for transgenesis, resulting in 25-50% germline transmission in F0-injected embryos. Mosimann was also interested in improving the generation of Cre/lox transgenes, which is notoriously difficult in zebrafish. Once again, the pIGLET fish were able to faithfully recapitulate previous loxP Switch transgenic lines with ease. This jumps a huge hurdle for the zebrafish community by allowing for more predictable creation of Cre/lox lines. Finally, the authors sought to determine if these lines were viable for the study of regulatory elements. “Our biggest problem was always regulatory element discovery and testing. […] With Tol2, you can screen yourself to death finding which [regulatory transgenic founders] have the right expression, patterns, levels and everything.” explained Mosimann. After creating both Tol2 and pIGLET versions of an enhancer deletion reporter model and a disease-associated noncoding variant enhancer reporter, the pIGLET models displayed expression that was consistent with, or even more complete than previous literature and in-house Tol2 models. “Having this streamlined [technique], with always having the same site, […] all of a sudden we can now do quantitative and qualitative comparisons of gene regulatory elements.” says Mosimann.
However, this tool does come with limitations. phiC31 incorporates the entire attB vector into the genome, which can increase transgene sizes. This can lead to higher rates of random background integration, which occurred in ~8% of pIGLET germline-transmission events in the study. However, the authors note that this predominantly occurs when plasmids are ~14kb or greater, which can be minimized by using minicircle plasmids1,13. Despite these limits, the pIGLET lines provide an extremely useful tool for efficient, reproducible transgenesis. The benefits also mean that multiple labs can accurately recreate the same transgenic lines with the necessary reagents and pIGLET fish, which the Mosimann team have already provided to over 60 labs. Overall, this tool is a huge boon for the zebrafish community. Both Lalonde and Mosimann encourage fellow researchers to reach out to procure the pIGLET lines and for any help with the transgenesis process. “It’s good to see that people are actively looking for these tools and getting them.” said Mosimann. “The greatest thing is when we see good news that its working!” added Lalonde.
References
1. Lalonde, R. L., Wells, H. H., Kemmler, C. L., Nieuwenhuize, S., Lerma, R., Burger, A., & Mosimann, C. (2024). pIGLET: Safe harbor landing sites for reproducible and efficient transgenesis in zebrafish. Science Advances, 10(23), eadn6603. https://doi.org/10.1126/sciadv.adn6603
2. Felker, A., & Mosimann, C. (2016). Contemporary zebrafish transgenesis with Tol2 and application for Cre/lox recombination experiments. In Methods in Cell Biology (Vol. 135, pp. 219–244). Elsevier. https://doi.org/10.1016/bs.mcb.2016.01.009
3. Soroldoni, D., Hogan, B. M., & Oates, A. C. (2009). Simple and Efficient Transgenesis with Meganuclease Constructs in Zebrafish. In G. J. Lieschke, A. C. Oates, & K. Kawakami (Eds.), Zebrafish (Vol. 546, pp. 117–130). Humana Press. https://doi.org/10.1007/978-1-60327-977-2_8
4. Albadri, S., Del Bene, F., & Revenu, C. (2017). Genome editing using CRISPR/Cas9-based knock-in approaches in zebrafish. Methods, 121–122, 77–85. https://doi.org/10.1016/j.ymeth.2017.03.005
5. Hu, G., Goll, M. G., & Fisher, S. (2011). ΦC31 Integrase Mediates Efficient Cassette Exchange in the Zebrafish Germline. Developmental Dynamics, 240(9), 2101–2107. https://doi.org/10.1002/dvdy.22699
6. Thorpe, H. M., & Smith, M. C. M. (1998). In vitro site-specific integration of bacteriophage DNA catalyzed by a recombinase of the resolvase/invertase family. Proceedings of the National Academy of Sciences, 95(10), 5505–5510. https://doi.org/10.1073/pnas.95.10.5505
7. Thorpe, H. M., Wilson, S. E., & Smith, M. C. M. (2000). Control of directionality in the site‐specific recombination system of the Streptomyces phage φC31. Molecular Microbiology, 38(2), 232–241. https://doi.org/10.1046/j.1365-2958.2000.02142.x
8. Knapp, J.-M., Chung, P., & Simpson, J. H. (2015). Generating Customized Transgene Landing Sites and Multi-Transgene Arrays in Drosophila Using phiC31 Integrase. Genetics, 199(4), 919–934. https://doi.org/10.1534/genetics.114.173187
9. Chen, C., Krohn, J., Bhattacharya, S., & Davies, B. (2011). A Comparison of Exogenous Promoter Activity at the ROSA26 Locus Using a PhiC31 Integrase Mediated Cassette Exchange Approach in Mouse ES Cells. PLoS ONE, 6(8), e23376. https://doi.org/10.1371/journal.pone.0023376
10. Mosimann, C., Puller, A., Lawson, K. L., Tschopp, P., Amsterdam, A., & Zon, L. I. (2013). Site‐directed zebrafish transgenesis into single landing sites with the phiC31 integrase system. Developmental Dynamics, 242(8), 949–963. https://doi.org/10.1002/dvdy.23989
11. Lister, J. A. (2010). Transgene excision in zebrafish using the phiC31 integrase. Genesis, 48(2), 137–143. https://doi.org/10.1002/dvg.20595
12. Roberts, J. A., Miguel-Escalada, I., Slovik, K. J., Walsh, K. T., Hadzhiev, Y., Sanges, R., Stupka, E., Marsh, E. K., Balciuniene, J., Balciunas, D., & Müller, F. (2014). Targeted transgene integration overcomes variability of position effects in zebrafish. Development, 141(3), 715–724. https://doi.org/10.1242/dev.100347
13. Shankar, R., Schmeer, M., & Schleef, M. (2017). Minicircles: Next-generation gene vectors. Cell and Gene Therapy Insights, 3(2), 285–300. https://doi.org/10.18609/cgti.2017.020
About the Authors

Dr. Robert Lalonde, the first author of this paper, is a K99-supported Postdoctoral Research Fellow in Dr. Christian Mosimann’s laboratory at the University of Colorado School of Medicine in the Department of Pediatrics, Section of Developmental Biology. His key research interests using zebrafish as principal model include the development and evolution of lateral plate mesoderm and how this development effects adult organ systems. He also has particular interest in the creation of transgenic lines and Cre/lox applications in the zebrafish system.
X - @Rob_Lalonde89
Dr. Christian Mosimann, last author of this paper, is a seasoned principal investigator. He holds the Johnson Chair and is an associate professor at the University of Colorado School of Medicine in the Department of Pediatrics, Section of Developmental Biology. His lab uses zebrafish as model to learn more about cell fate determination in development and disease. His key interests are mesodermal cell fates, lateral plate mesoderm emergence, and cardiovascular development & congenital heart disease. His lab’s research also continues to utilize and develops transgenic and genome editing techniques.
X - @chrmosimann
BlueSky - @chrmosimann.bsky.social
About the Science Spotlight Writer

Jennifer Fiene is a Master’s student in Dr. Jason Berman’s laboratory at the University of Ottawa & CHEO Research Institute in Canada. Her research focuses on using zebrafish to model the genetic cancer predisposition disorder Li-Fraumeni syndrome. She is also very passionate about science communication and increasing science accessibility within and outside of scientific communities.
LinkedIn - Jennifer Fiene (www.linkedin.com/in/jennifer-fiene-9b7331274)