Congratulations to the IZFS 2023 Chi-Bin Chien & George Streisinger Award Winners!
2023 Chi-Bin Chien Award Winner: Peter Fabian, PhD, Masaryk University,
Brno, Czech Republic
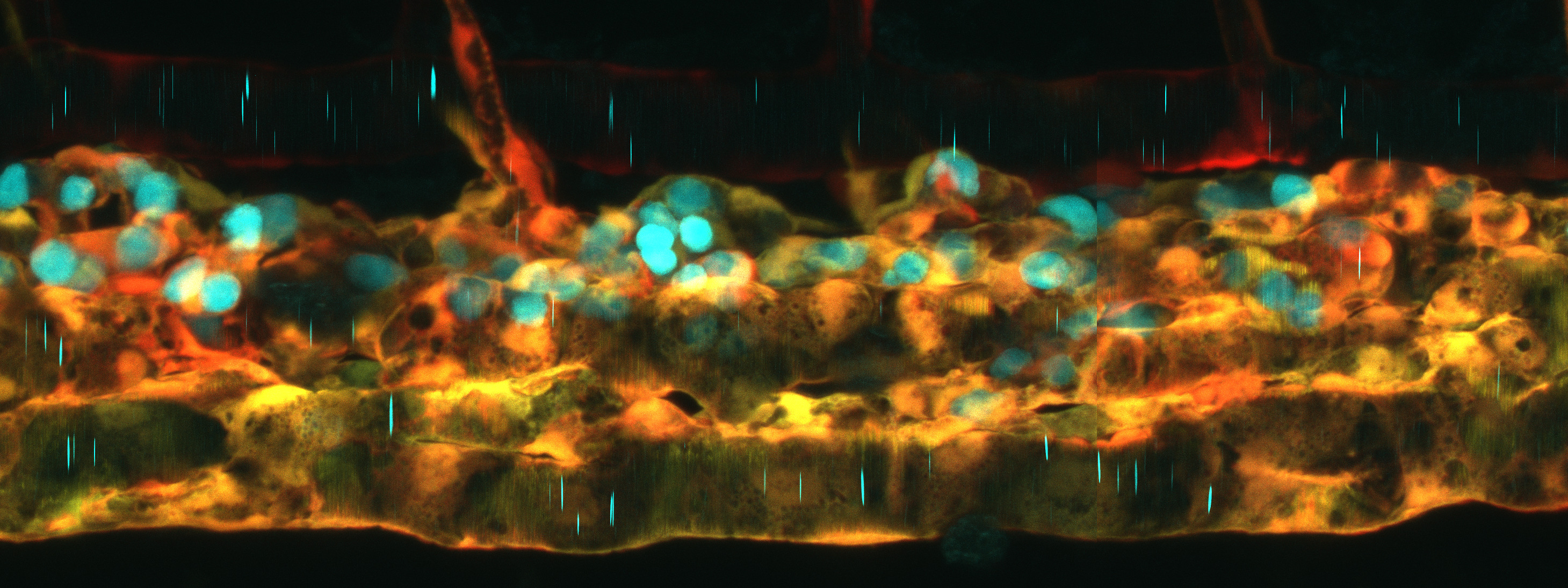
Dr. Peter Fabian is a researcher in the field of developmental biology with a focus on understanding the fundamental processes that underlie the development and maintenance of vertebrate organs, particularly their relevance to human health. In the fall of 2022, he started working as an assistant professor at the Department of Experimental Biology at Masaryk University. Dr. Fabian began his work in the field of developmental biology during his Ph.D. studies in Zbynek Kozmik's laboratory at Charles University in Prague, where he also studied developmental and evolutionary biology with Robert Cerny. He acquired a skillset in zebrafish genetics, in vivo imaging, and single-cell approaches through his postdoctoral studies in Gage Crump's lab at the University of Southern California. Today, he sees as a priority to share this knowledge to the scientific community both locally (in the Czech Republic) and globally.
Click here to visit his lab. Click here to visit his Twitter
2023 George Streisinger Award Winner: John Postlethwait, PhD, University of Oregon,
Eugene, Oregon, USA

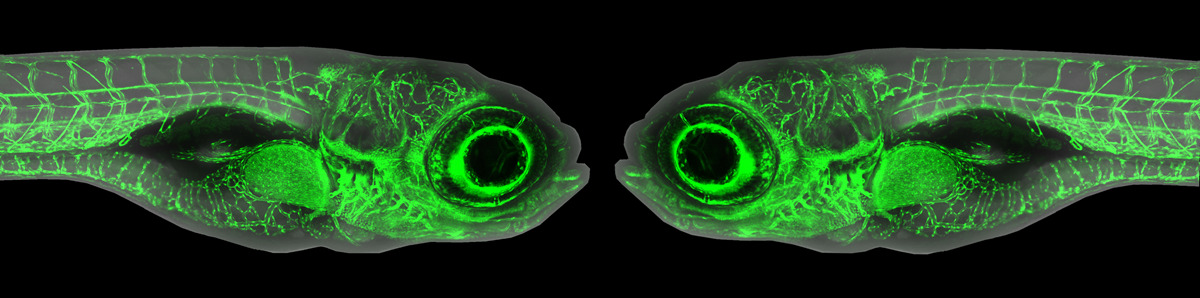
I trained initially in Drosophila developmental genetics and later switched to zebrafish developmental genetics and genomics to capitalize on a vertebrate model. We showed that the lineage of teleost fish, including zebrafish, experienced a whole genome duplication event and provided an understanding of how the zebrafish genome relates to the human genome. We have also developed several zebrafish models for human disease, including Fanconi anemia, craniofacial diseases, and reproductive diseases. For Fanconi anemia, we showed that zebrafish has, unexpectedly, a single ortholog of each of the (now) 19 human FA genes, created ENU-induced mutants in five zebrafish FA genes that mimic the double-strand break induced chromosome damage, hypogonadal, and cancer phenotypes of human patients and ran a small molecule screen that identified a compound that rescues the DNA-damage phenotype. We created mutant models for several craniofacial diseases, including microRNA140 that spurred physicians to identify a mutation in microRNA140 in several non-syndromic cleft palate families. We used forward mutation screens to find mutants in Fraser syndrome, which mimics the fluctuating asymmetry displayed by human patients. For reproductive diseases, we used genome editing to make mutants in three different genes that mimic polycystic ovarian syndrome, the most common problem when couples visit fertility clinics. Experience in connecting the human and zebrafish genome and in creating zebrafish disease models prepares me for my role in this project.
Click here to visit his lab.