Science Spotlight - single-cell transcriptomics
An integrated view of single-cell transcriptomics with spatially resolved imaging
By Cynthia Ufuoma Adjekukor,
PhD candidate in Biochemistry and Molecular Biology,
Childs lab, University of Calgary, Canada.
Since ancient times, biologists have sought to comprehend how cells package into tissues, organs, and finally, an organism. Although every cell has the same DNA, each organ is unique due to the specific gene sets expressed by each cell at distinct times and locations. This question has accelerated the advancement of technology to gain a deeper understanding of gene expression and regulation at the cellular level. The zebrafish has emerged as a valuable model organism for studying embryo and organ development. In the mid-1990s, before the zebrafish genome project and RNA sequencing in the 2000s, tools like microarrays were employed to study gene expression data. These were crucial in discovering new genes, mapping them, modeling human diseases, and understanding transcriptome levels and changes. However, while scientists could tell a gene was expressed at a specific time or condition, they still lacked precise information on the gene of interest's spatial expression.
Identifying ways to resolve this became the forefront of research in the early and mid-2010s with the invention of single-cell RNA (scRNAseq) and ATAC (scATACseq) sequencing to understand transcriptomic and epigenetic regulation at the cellular level. Now scientists can tell if a gene is expressed in a specific cell type and what epigenetic modification controls this. Given how beneficial these datasets became in understanding cellular gene expression and heterogeneity, discovering new cell populations, and uncovering complex biological processes and possible disease biomarkers, scientists began to create whole-organism cell atlases to serve as a resource for future researchers. The Zebrafish community has evolved from the breakthrough single-cell transcriptomics atlas that covers 24hpf whole embryonic development (1) to a more comprehensive atlas like daniocell (2,3) that covers 3.3 – 120hpf zebrafish embryonic development or zscape (4,5) that covers 18 – 96hpf wild type vs temperature-perturbed embryonic development.
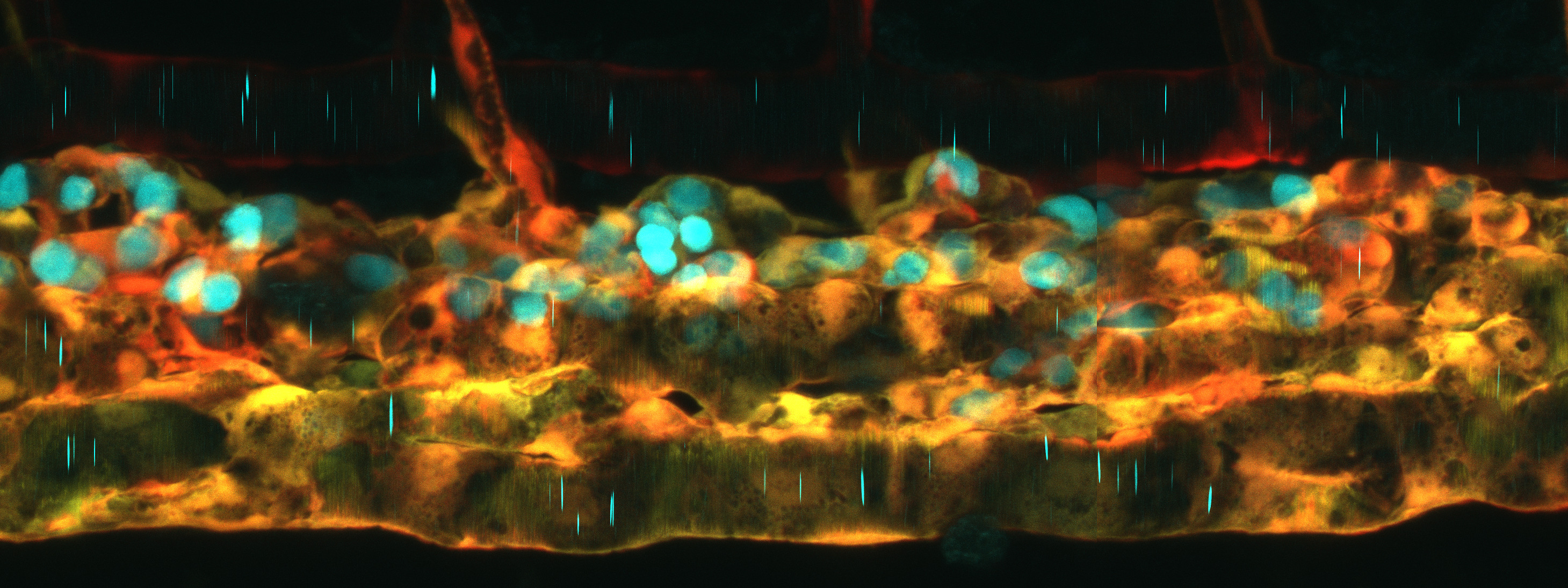
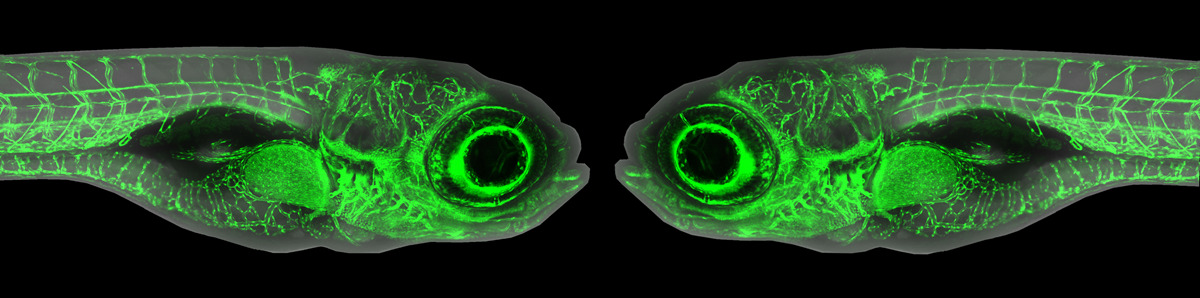
A new resource tool, Zebrahub, offers single-cell transcriptomics with spatially resolved images. Zebrahub integrates transcriptomics (scRNAseq) data across 10 developmental stages (10hpf, 12hpf, 14hpf, 16hpf, 19hpf, 1dpf, 2dpf, 3dpf, 5dpf, 10dpf) with light sheet microscopy to enable in silico fate mapping of cell lineages during development (6). Utilizing this integrated toolset, Dr Loic Royer’s team at the Chan Zuckerberg Biohub and collaborators studied the development of the tail and of mesodermally-derived organs. They achieved this by using RNA velocity to identify genes expressed in progenitors in the tail bud and lateral plate mesoderm (LMP) and validating their expression by RNA in situ. Then, they performed in silico fate mapping of cells estimated to lie in the corresponding expression domains by marking them in a high-resolution light-sheet timelapse of early embryo development (7). Using an in-silico fate mapping plugin they followed the fates of all the marked cells in the virtual embryo. All of the materials for this analysis have been made available at zebrahub. Authors Dr Merlin Lange and Dr. Loic Royer said, “Zebrahub is a flagship resource of the past 5 – 7 years. We trusted in our abilities, we had confidence in our science, and we faced challenges, but we persevered because we know how valuable this tool will be to the Zebrafish research community”.
After gastrulation has generated the three tissue layers, the tailbud continues to house bipotential neuromesodermal progenitors (NMPs) as the tail grows out. Lange et al. used their in-silico fate mapping method to follow NMPs in silico. In early segmentation stages, NMP cells can give rise to mesoderm and neural tissues. However, after 14hpf, they become restricted to mesodermal cell fates. This fate restriction correlates with changes in tissue architecture: cells become more coherent as they become more fate restricted. Lange et al. also followed LPM progenitors, which are known to give rise to the hematopoietic-endothelial lineages. However, they observed that during early somitogenesis, early LPM progeny can also contribute to the pronephric lineages.
Zebrahub will serve as a benchmark tool for researchers interested in exploring gene expression profiles in specific tissue locations during early development and organogenesis. However, most cell lineage answers from scRNAseq rely heavily on the RNA velocity map flow. Precaution needs to be taken when using this as the velocity map is based on spliced vs unspliced transcripts and can yield wrong cell expression trajectories in genes/cells with low splicing rate. Also, it is possible to over/under-populate gene trajectories in specific cell populations during RNA velocity map creation. This is why the light sheet images were created to supplement the transcriptomic data to improve our confidence in fate mapping studies. However, image validation is currently limited to only 10 – 24hpf embryonic development and therefore misses most of organogenesis. Nevertheless, zebrahub is a step in the right direction in understanding cell differentiation and tissue dynamics, and with improvements in expanding imaging time points, this will provide more robust in silico cell tracking, making the potential for new knowledge gained using this publicly available resource limitless. Currently, the scATACseq data is being set up to integrate gene expression modifications to existing scRNASeq and imaging data across six developmental time points (10hpf, 12hpf, 14hpf, 16hpf, 19hpf, 24hpf) (8). These tools can further enhance the utility of Zebrafish to investigate gene expression and regulation at the spatial tissue level, providing information on cell behavior, differentiation, lineage, and organization into specialized tissues.
The authors are strongly motivated to make their data accessible to the community: “We view science as an art, so we are constantly developing, especially in the user accessibility area. We plan to make an interactive web design for research analysis and have more plans on the technical aspects for the future. Our biggest satisfaction will come from the research community using our tool to answer their biological questions”.
References
1. Wagner DE, Weinreb C, Collins ZM, Briggs JA, Megason SG, Klein AM. Single-cell mapping of gene expression landscapes and lineage in the zebrafish embryo. Science. 2018 Jun 1;360(6392):981–7.
2. Farrell JA, Wang Y, Riesenfeld SJ, Shekhar K, Regev A, Schier AF. Single-cell reconstruction of developmental trajectories during zebrafish embryogenesis. Science. 2018 Jun 1;360(6392):eaar3131.
3. Sur A, Wang Y, Capar P, Margolin G, Prochaska MK, Farrell JA. Single-cell analysis of shared signatures and transcriptional diversity during zebrafish development. Developmental Cell. 2023 Dec 18;58(24):3028-3047.e12.
4. Saunders LM, Srivatsan SR, Duran M, Dorrity MW, Ewing B, Linbo TH, et al. Embryo-scale reverse genetics at single-cell resolution. Nature. 2023 Nov 1;623(7988):782–91.
5. Dorrity MW, Saunders LM, Duran M, Srivatsan SR, Barkan E, Jackson DL, et al. Proteostasis governs differential temperature sensitivity across embryonic cell types. Cell. 2023 Nov 9;186(23):5015-5027.e12.
6. Lange M, Granados A, VijayKumar S, Bragantini J, Ancheta S, Kim YJ, et al. A multimodal zebrafish developmental atlas reveals the state-transition dynamics of late-vertebrate pluripotent axial progenitors. Cell. 2024 Nov 14;187(23):6742-6759.e17.
7. Yang B, Lange M, Millett-Sikking A, Zhao X, Bragantini J, VijayKumar S, et al. DaXi—high-resolution, large imaging volume and multi-view single-objective light-sheet microscopy. Nature Methods. 2022 Apr 1;19(4):461–9.
8. Kim YJ, Vijay Kumar S, Iovino B, Granados A, Ancheta S, Zhao X, et al. Zebrahub-Multiome: Uncovering Gene Regulatory Network Dynamics During Zebrafish Embryogenesis. bioRxiv. 2024 Jan 1;2024.10.18.618987.
About the Science Spotlight Writer and Authors
 Dr. Loic Royer holds an Engineering degree from CY Tech France, a master’s degree specialization in Artificial Intelligence and a PhD in Bioinformatics from Technische Universität Dresden Germany. For his post-doc he joined Dr Eugene Myers’ group at HMMI’s Janelia Farms Research Campus where, in collaboration with Dr Philipp Keller, he developed the first adaptive multi-view light sheet microscope. As a multidisciplinary group leader at the Chan Zuckerberg (CZ) Biohub Dr. Royer is interested in understanding vertebrate development using microscopy and bioinformatics tools.
Dr. Loic Royer holds an Engineering degree from CY Tech France, a master’s degree specialization in Artificial Intelligence and a PhD in Bioinformatics from Technische Universität Dresden Germany. For his post-doc he joined Dr Eugene Myers’ group at HMMI’s Janelia Farms Research Campus where, in collaboration with Dr Philipp Keller, he developed the first adaptive multi-view light sheet microscope. As a multidisciplinary group leader at the Chan Zuckerberg (CZ) Biohub Dr. Royer is interested in understanding vertebrate development using microscopy and bioinformatics tools.
 Dr. Merlin Lange received his PhD in neuroscience from the CNRS Institut des Neurosciences Paris-Saclay under the supervision of Dr Laure Bally-Cuif. There, he worked on zebrafish models of neurodevelopmental disorders. He received the Japanese special researcher postdoctoral program at the RIKEN Center for Brain Science to study zebrafish neural networks in Dr Hitoshi Okamoto's group. With a team of computer scientists, software engineers, optical engineers, bioinformaticians at the CZ biohub, Dr. Lange works to answer complex developmental questions such as how a single cell becomes an organism.
Dr. Merlin Lange received his PhD in neuroscience from the CNRS Institut des Neurosciences Paris-Saclay under the supervision of Dr Laure Bally-Cuif. There, he worked on zebrafish models of neurodevelopmental disorders. He received the Japanese special researcher postdoctoral program at the RIKEN Center for Brain Science to study zebrafish neural networks in Dr Hitoshi Okamoto's group. With a team of computer scientists, software engineers, optical engineers, bioinformaticians at the CZ biohub, Dr. Lange works to answer complex developmental questions such as how a single cell becomes an organism.

Dr. Cynthia Ufuoma Adjekukor is currently a PhD candidate in Biochemistry and Molecular Biology at the University of Calgary under the supervision of Dr Sarah Childs. She is a recipient of the Eyes High Doctoral Recruitment scholarship. She studies the connection between mural cell populations and the brain endothelium in angiogenesis, vascular stability, and cerebral small vessel diseases. Outside the lab, she participates in numerous educational programs for grade 5 - 10 students. She was the Calgary library math quest instructor and a STEM professional in Letters to Pre-Scientist (2021-2023). Besides academics, she enjoys watching and critiquing films, particularly those from Nollywood, the Nigerian film industry. She intends to continue her academic journey by obtaining a postdoctoral position after completing her PhD studies.